import numpy as np
import matplotlib.pyplot as plt
from scipy.fft import fft, ifft
from scipy.io import loadmatChapter 5
Chapter 5
Analyzing Neural Time Series Data
Python code for Chapter 5 – converted from original Matlab by AE Studio (and ChatGPT)
Original Matlab code by Mike X Cohen
This code accompanies the book, titled “Analyzing Neural Time Series Data” (MIT Press).
Using the code without following the book may lead to confusion, incorrect data analyses, and misinterpretations of results.
Mike X Cohen and AE Studio assume no responsibility for inappropriate or incorrect use of this code.
Import necessary libraries
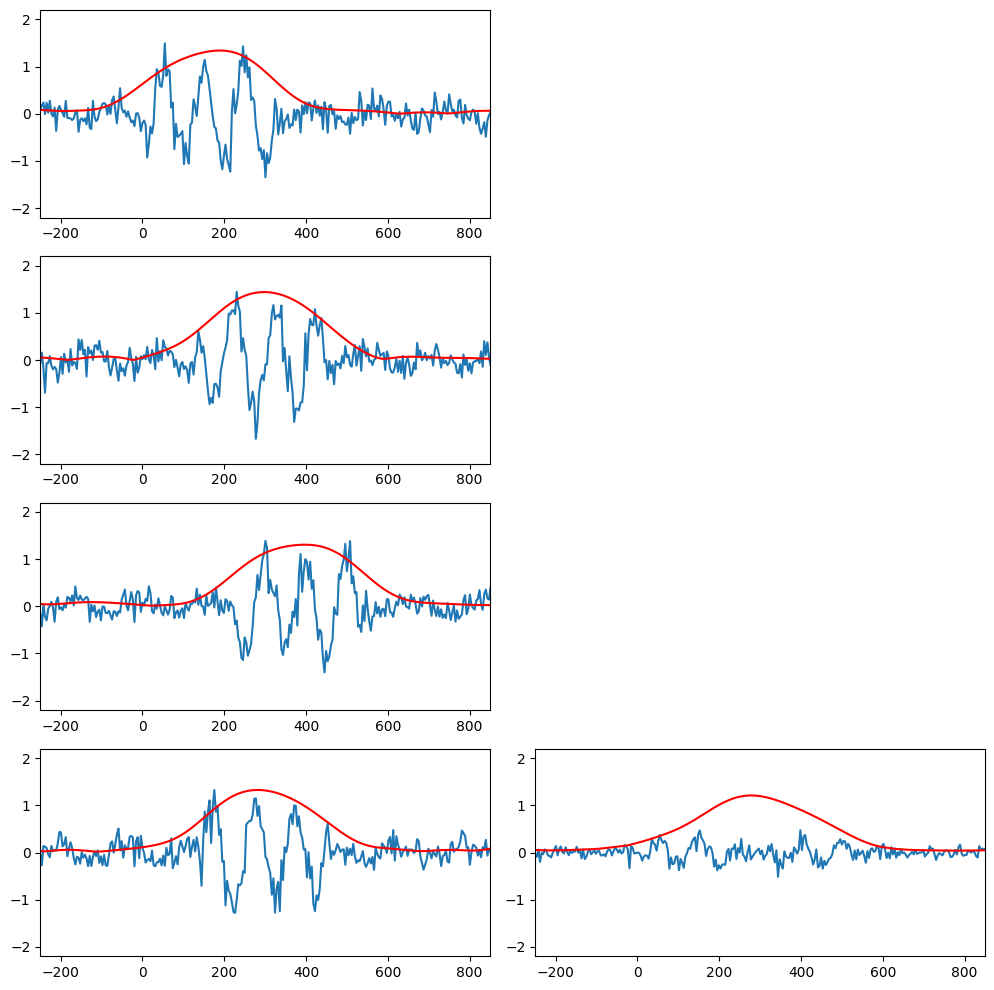
Time-locked and non-phase-locked analysis
# Load sample data
EEG = loadmat('../data/sampleEEGdata.mat')['EEG'][0, 0]
nTrials = 4
data = np.zeros((nTrials, EEG['pnts'][0][0]))
# Wavelet parameters
wavetime = np.arange(-1, 1 + 1/EEG['srate'][0][0], 1/EEG['srate'][0][0])
n_conv = len(wavetime) + EEG['pnts'][0][0] - 1
waveletfft = fft(np.exp(2 * 1j * np.pi * 10 * wavetime) * np.exp(-wavetime ** 2 / (2 * (5 / (2 * np.pi * 10)) ** 2)) / 10, n_conv)
data10hz = np.zeros((nTrials, EEG['pnts'][0][0]))
plt.figure(figsize=(15, 10))
for triali in range(nTrials):
# Create single trial ERP as sine wave plus noise
data[triali, :] = 0.15 * np.sin(2 * np.pi * 6 * EEG['times'][0] / 1000 + 2 * np.pi * (triali + 1) / nTrials - np.pi) + np.random.randn(EEG['pnts'][0][0]) / 6
# Add non-phase-locked stimulus potential
data[triali, 259:360] = data[triali, 259:360] + np.sin(2 * np.pi * 10 * EEG['times'][0][259:360] / 1000 + 2 * np.pi * (triali + 1) / nTrials - np.pi) + np.random.randn(101) / 5
# Convolve with 10Hz wavelet
convolution_result_fft = ifft(waveletfft * fft(data[triali, :], n_conv)) * np.sqrt(5 / (2 * np.pi * 10))
convolution_result_fft = convolution_result_fft[int(np.floor(len(wavetime) / 2)) : -int(np.floor(len(wavetime) / 2))]
data10hz[triali, :] = np.abs(convolution_result_fft)*2
# Plot single trials
plt.subplot(nTrials, 3, (triali) * 3 + 1)
plt.plot(EEG['times'][0], data[triali, :])
plt.plot(EEG['times'][0], data10hz[triali, :], 'r')
plt.xlim([-250, 850])
plt.ylim([-2.2, 2.2])
# Plot the mean over trials
plt.subplot(nTrials, 3, (triali) * 3 + 2)
plt.plot(EEG['times'][0], np.mean(data, axis=0))
plt.plot(EEG['times'][0], np.mean(data10hz, axis=0), 'r')
plt.xlim([-250, 850])
plt.ylim([-2.2, 2.2])
plt.tight_layout()
plt.show()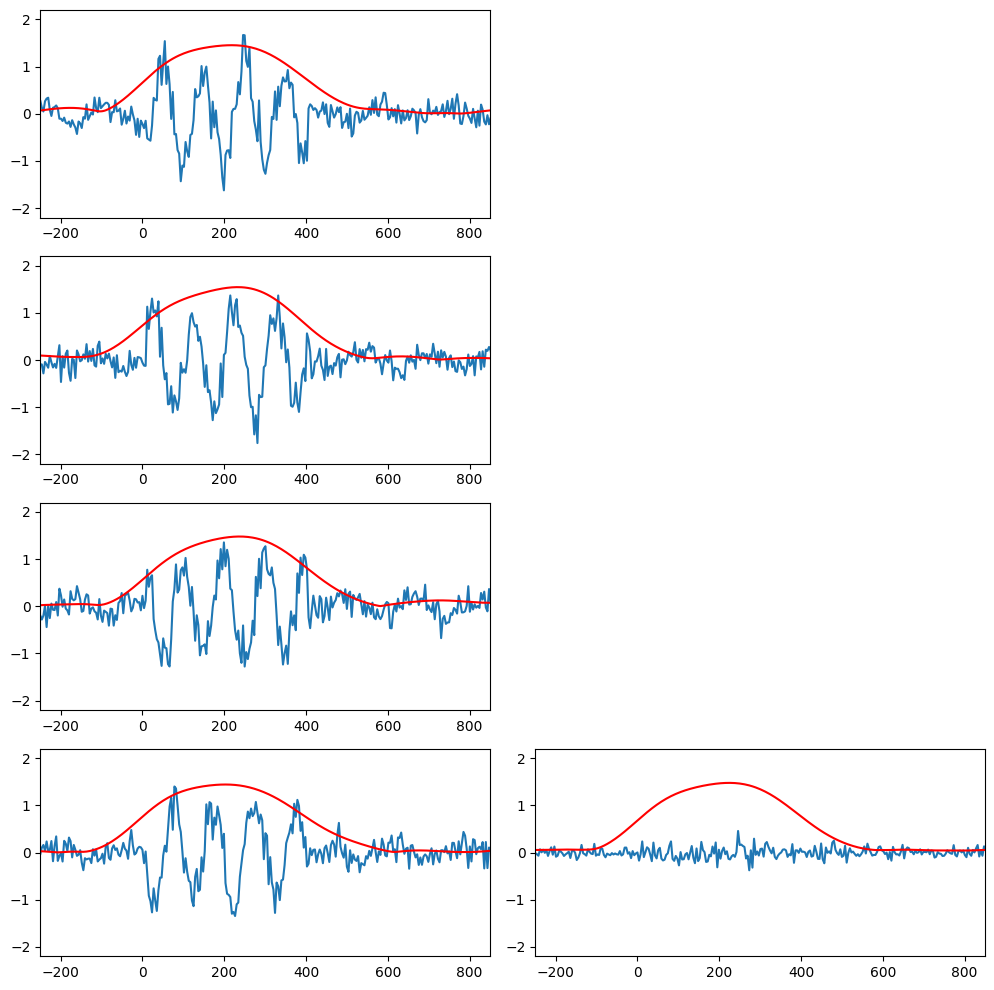
Time-locked and phase-locked analysis
nTrials = 4
data = np.zeros((nTrials, EEG['pnts'][0][0]))
# Wavelet parameters
wavetime = np.arange(-1, 1 + 1/EEG['srate'][0][0], 1/EEG['srate'][0][0])
n_conv = len(wavetime) + EEG['pnts'][0][0] - 1
waveletfft = fft(np.exp(2 * 1j * np.pi * 10 * wavetime) * np.exp(-wavetime ** 2 / (2 * (5 / (2 * np.pi * 10)) ** 2)) / 10, n_conv)
data10hz = np.zeros((nTrials, EEG['pnts'][0][0]))
plt.figure(figsize=(15, 10))
for triali in range(nTrials):
# Create single trial ERP as sine wave plus noise
data[triali, :] = 0.15 * np.sin(2 * np.pi * 6 * EEG['times'][0] / 1000 + 2 * np.pi * (triali + 1) / nTrials - np.pi) + np.random.randn(EEG['pnts'][0][0]) / 6
# Add phase-locked stimulus potential
data[triali, 259:360] = data[triali, 259:360] + np.sin(2 * np.pi * 10 * EEG['times'][0][259:360] / 1000) + np.random.randn(101) / 5
# Convolve with 10Hz wavelet
convolution_result_fft = ifft(waveletfft * fft(data[triali, :], n_conv)) * np.sqrt(5 / (2 * np.pi * 10))
convolution_result_fft = convolution_result_fft[int(np.floor(len(wavetime) / 2)) : -int(np.floor(len(wavetime) / 2))]
data10hz[triali, :] = np.abs(convolution_result_fft)*2
# Plot single trials
plt.subplot(nTrials, 3, (triali) * 3 + 1)
plt.plot(EEG['times'][0], data[triali, :])
plt.plot(EEG['times'][0], data10hz[triali, :], 'r')
plt.xlim([-250, 850])
plt.ylim([-2.2, 2.2])
# Plot the mean over trials
plt.subplot(nTrials, 3, (triali) * 3 + 2)
plt.plot(EEG['times'][0], np.mean(data, axis=0))
plt.plot(EEG['times'][0], np.mean(data10hz, axis=0), 'r')
plt.xlim([-250, 850])
plt.ylim([-2.2, 2.2])
plt.tight_layout()
plt.show()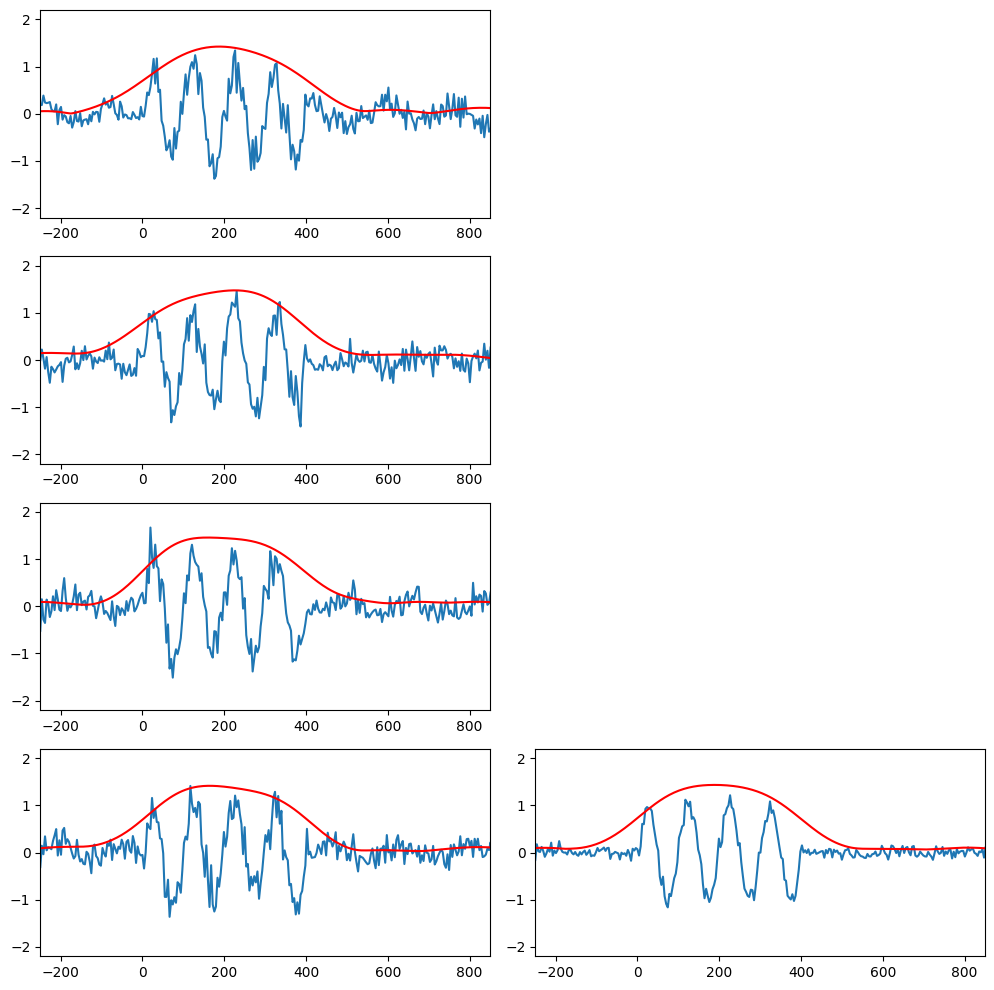
Non-time-locked and phase-locked analysis
nTrials = 4
data = np.zeros((nTrials, EEG['pnts'][0][0]))
# Wavelet parameters
wavetime = np.arange(-1, 1 + 1/EEG['srate'][0][0], 1/EEG['srate'][0][0])
n_conv = len(wavetime) + EEG['pnts'][0][0] - 1
waveletfft = fft(np.exp(2 * 1j * np.pi * 10 * wavetime) * np.exp(-wavetime ** 2 / (2 * (5 / (2 * np.pi * 10)) ** 2)) / 10, n_conv)
data10hz = np.zeros((nTrials, EEG['pnts'][0][0]))
plt.figure(figsize=(15, 10))
for triali in range(nTrials):
# Create single trial ERP as sine wave plus noise
data[triali, :] = 0.15 * np.sin(2 * np.pi * 6 * EEG['times'][0] / 1000 + 2 * np.pi * (triali + 1) / nTrials - np.pi) + np.random.randn(EEG['pnts'][0][0]) / 6
# Add non-time-locked, phase-locked stimulus potential
eventtime = np.random.permutation(80) + 240
eventtime = range(eventtime[0], eventtime[0] + 81)
data[triali, eventtime] = data[triali, eventtime] + np.sin(2 * np.pi * 10 * EEG['times'][0][eventtime] / 1000 + 2 * np.pi * (triali + 1) / nTrials - np.pi) + np.random.randn(len(eventtime)) / 5
# Convolve with 10Hz wavelet
convolution_result_fft = ifft(waveletfft * fft(data[triali, :], n_conv)) * np.sqrt(5 / (2 * np.pi * 10))
convolution_result_fft = convolution_result_fft[int(np.floor(len(wavetime) / 2)) : -int(np.floor(len(wavetime) / 2))]
data10hz[triali, :] = np.abs(convolution_result_fft)*2
# Plot single trials
plt.subplot(nTrials, 3, (triali) * 3 + 1)
plt.plot(EEG['times'][0], data[triali, :])
plt.plot(EEG['times'][0], data10hz[triali, :], 'r')
plt.xlim([-250, 850])
plt.ylim([-2.2, 2.2])
# Plot the mean over trials
plt.subplot(nTrials, 3, (triali) * 3 + 2)
plt.plot(EEG['times'][0], np.mean(data, axis=0))
plt.plot(EEG['times'][0], np.mean(data10hz, axis=0), 'r')
plt.xlim([-250, 850])
plt.ylim([-2.2, 2.2])
plt.tight_layout()
plt.show()